
If this is your first time reading my newsletter: I am thrilled that you decided to give it try!
If this is not your first time: I’m glad you’re still here!
We’ve got a few things to go through in this week’s edition.
However, before we get into the cool stuff, that is showcasing useful functionality and interesting use cases, I feel it would behoove me to lay down some of the foundational things you’ll need to do to get you started in R, should you be interested.
If you’re already an R user: great, you can skip through the next couple sections. If you’ve never set up and used R though, you may want to stick around for a few pointers. It won’t be a step-by-step tutorial by any means, but I will summarize key actions you need to take as well as share with you a few helpful resources and code for this week’s edition.
Installing R and RStudio (skip this if you’re already an R user)
In order to use R, you need to install both R, the programming language itself, as well as a user interface that allows you to interact with R. Well, that interface is RStudio, developed by Posit (formerly known as RStudio). The analogy is that R is the car’s engine and RStudio is the car’s dashboard, if you will, and you need both to be able to drive your car.
The good news is installing both R and RStudio is pretty quick and simple. There are plenty of excellent resources on how to do this. Follow the instructions in this one, and you should be good to go. If you prefer video instructions, check out this YouTube video.
Launching and setting up RStudio (skip this if you’re already an R user)
Once you’ve installed RStudio, you can just go ahead and simply launch it.
There are a few important elements that you need to be aware of, at least for now:
- The Console
- The Source Pane
- The Environment pane
- The Plots pane
More on the layout and basic setup here.
Installing and loading packages (skip this if you’re already an R user)
R as a programming language has a lot of functionality out of the box, but in order to access some of the best pieces of functionality, you’ll need to install and load what’s called packages. Packages can house both functionality in the form of functions (more on how to execute a function in a little bit) and data.
Installing packages might sound like a tedious task, but in reality it’s not. It’s a one-time thing (for the most part) and can easily be done by calling install.packages(”package_name”), for example install.packages(”tidyverse”).
Once you’ve installed a package, you’ll need to load it every time you want to use it by calling library(package_name), for example library(tidyverse).
# 0. INSTALLING PACKAGES ----
# This is a one-time thing
# 0.1 Either installing one package at a time ----
install.packages("tidyverse")
install.packages("sherlock")
install.packages("openxlsx")
# 0.2 or Installing multiple packages through concatenation ----
install.packages(c("tidyverse", "sherlock", 'openxlsx'))
# 0.3 or Assigning as a variable and calling it ----
packages <- c("tidyverse", "sherlock", 'openxlsx')
install.packages(packages)
# 1. LOADING PACKAGES (LIBRARIES) ---
# You'll need to load packages every time you intend to use them
library(tidyverse)
library(sherlock)
library(openxlsx)Reading data into memory a.k.a opening files from spreadsheets
While there are many ways to import data into RStudio’s environment (including from databases, from the cloud, from Google Drive etc.), chances are a good portion of the time you will be working with data coming straight from good ol’ Excel or a text file. Let’s explore this a little bit.
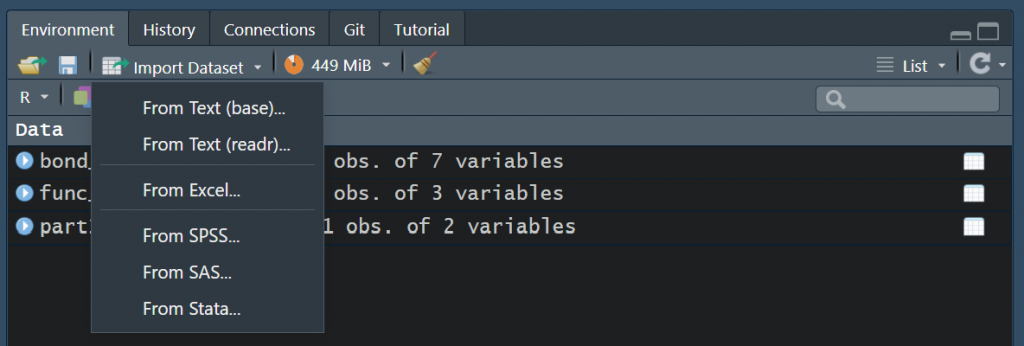
As a newcomer to R not being familiar with doing stuff from the command line, you might, very rightfully, ask, “Where’s my Open File button?”. The good news is, there is one. Just go to the Environment tab on the right hand side and click Import Dataset, and you will be presented with a few options, the most notable of which being the one that says From Excel… You can also import .csv and .txt files by clicking From Text (reader).
If you want to get an idea how to read in a file programmatically, take a look at the below script. The load_file() function from the sherlock package is an all-in-one function to read in tabular files of various types.
# 2. READING FILE INTO MEMORY ----
# 2.1 From RStudio: Go to Environment -> Import Dataset ----
# 2.2 Programmatically: .XLSX FILES USING OPENXLSX::READ.XLSX() ----
read.xlsx(xlsxFile = "../../00_data/bond_strength.xlsx")
# 2.3 Programmatically: .CSV FILES USING READR::READ_CSV() ----
read_csv(file = "../../00_data/experiment_2.csv")
# 2.4 Programmatically: .TXT FILES USING READR::READ_TABLE() ----
read_table(file = "../../00_data/experiment_3_data.txt")
# 2.4 Programmatically: ALL-IN-ONE USING SHERLOCK::LOAD_FILE() ----
load_file(path = "../../00_data/bond_strength.xlsx", filetype = ".xlsx")
load_file(path = "../../00_data/experiment_2.csv", filetype = ".csv")
load_file(path = "../../00_data/experiment_3_data.txt", filetype = ".txt")
Basic data exploration
Let’s go over different ways to do basic data exploration, that is, just examining your data just like you would if you were using say Excel.
Run the below code snippet to read in a dataset from one of my GitHub repositories and assign it to a variable called bond_strength. Notice that assignment is done using the ← operator (Alt + Dash).
# 3. BASIC DATA EXPLORATION ----
# 3.1 ASSIGNING TO A VARIABLE CALLED "BOND_STRENGTH" ----
# Run the below function call by selecting lines 50-52 and hitting Ctrl + Enter.
# Pulls file from GitHub and assigns it to a variable named bond_strength.
bond_strength <- load_file(
path = "https://raw.githubusercontent.com/gaboraszabo/datasets-for-sherlock/main/bond_strength_multivari.csv",
filetype = ".csv")You then have a few different ways to examine the dataset.
# 3.2 DISPLAYING FIRST TEN ROWS IN THE CONSOLE ----
# Simply run this
bond_strength
# 3.3 VIEW()
bond_strength %>% View()
View(bond_strength)
# 3.4 GLIMPSE()
bond_strength %>% glimpse()
glimpse(bond_strength)
# 3.5 HEAD(), TAIL()
bond_strength %>%
head()Notice the use of the pipe operator, %>% (Ctrl + Shift + M). This thing come in really handy as it chains multiple operations together, making your code much easier to read. You can chain as many operations together as you want, and I will discuss examples of that in later. In the meantime, you can take a look at this resource to learn more about pipes.
As a demostration of how pipes work, let’s take line 63 from the above snippet where we’ve chained the variable bond_strength and the glimpse() function together.
Here’s what it means:
take the variable (dataset) called bond_strength THEN run the glimpse() function on it
Alternatively, you could also just say glimpse(bond_strength), see line 64, and that would result in the same outcome.
Here’s a link to the above script if you would like to download it.
A simple plotting function
Once you’ve imported the dataset of interest and done some exploration, you may decide to do some sort of graphical analysis on it.
To do that, you could build your own graph from scratch, and doing so will make sense a lot of the time; certain packages such as ggplot2 will enable you to do just that and then some. I cannot emphasize enough how much one can accomplish with simply plotting the data (”Plot your data!” as visionary statistician Ellis Ott would say), and the functionality that comes with packages such as ggplot2, plotly and a few other packages is hard to beat—but you have to learn how to do it. And you should.
But for now, let’s just start with what I call a ready-made plotting function that’s also customizable enough.
We are going to plot the bond strength results from bond_strength dataset in what I call a Categorical Scatterplot (an Inidividual Value Plot in Minitab terminology). All we need to do is simply call the variable, pipe it into the draw_categorical_scatterplot() function, specify the main argument of the function and hit Ctrl + Enter to run it. Bam. Plot created.
bond_strength %>%
draw_categorical_scatterplot(y_var = `Bond Strength`)If you want to take it up a notch, you could specify some of the other arguments such as a grouping variable, whether or not you want the means of the groups plotted and whether or not to plot each group in a unique color, which would look something like this:
bond_strength %>%
draw_categorical_scatterplot(y_var = `Bond Strength`,
grouping_var_1 = `Bonding Station`,
plot_means = TRUE,
group_color = TRUE)Note that there is a lot more functionality built into the draw_categorical_scatterplot() function, and I would encourage you to experiment with it.
Now, one could build the same plot from scratch, but it would take a some time to customize it. With a few function calls though, you could do this:
bond_strength %>%
mutate(`Bonding Station` = `Bonding Station` %>% as_factor()) %>%
ggplot(aes(x = `Bonding Station`, y = `Bond Strength`, color = `Bonding Station`)) +
geom_point() +
stat_summary(fun = "mean", color = "black")Notice that this indeed was multiple lines of code and took multiple functions chained together to make a somewhat OK looking plot. It definitely does the trick, but it doesn’t look as pretty out of the box as a ready-made plotting function specifically designed for that purpose. And this is what can make using R relatively easy—you don’t always need to worry about creating something from scratch as the specific functionality you’re looking for, say a specific plot, has already been built by someone else and wrapped into a ready-made function, and using it will likely make your life much easier.
And to save you some time that would otherwise spend on finding those useful pieces of functionality, I will focus on showcasing ready-made functionality that hopefully will make your work easier and better.
That’s it for this week. Next week we will dive head first into an interesting use case!
Download the code for this week’s session here.
Thanks for reading, I will see you next week. Until then, keep exploring!




 Ask a question or send along a comment.
Please login to view and use the contact form.
Ask a question or send along a comment.
Please login to view and use the contact form.
Congratulations and thanks for introducing R to reliability engineers. There are several R-packages for survival analysis that could be useful [“survival”and “surv”], if users have life data.
Mark Felthauser and I translated my VBA software for nonparametric estimation of reliability functions, without life data, into R scripts. They include renewal processes where only cohort sizes and failure counts (periodic “ships” and “returns”) are known. Periodic failure counts for renewal processes don’t tell how many prior failures there were. The R community on http://www.LinkedIn,com didn’t believe reliability estimation could be done without life data, but maybe reliability engineers might be interested. GAAP requires statistically sufficient data.
Looking forward to more.
Larry, I am thrilled to hear you’re enjoying the series so far!
Reliability is definitely one of the areas where R has useful functionality. I haven’t used the “survival”and “surv” packages yet and would be interested in learning more about them myself.
In future editions of the newsletter I will try and cover topics and tools as it relates to reliability engineering.